Research Article | DOI: https://doi.org/10.58489/2836-2187/015
Toxicity of Diphenyltin Chloride on viable count of aerobic bacteria from River Benue.
1Department of Microbiology, Joseph Sarwuan Tarka University, Makurdi.
2Department of Chemistry, Joseph Sarwuan Tarka University, Makurdi
*Corresponding Author: Ichor T*
Citation: Ichor T, Ate M.L, Ebah E.E and Amua Q, Toxicity of Diphenyltin Chloride on the viable count of aerobic bacteria from River Benue. 2(2). DOI: 10.58489/2836-2187/015
Copyright: © 2023 Ichor T, this is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: 05 July 2023 | Accepted: 04 August 2023 | Published: 21 August 2023
Keywords: Diphenyltin Chloride, Organotin, Di-alkyl Organotins, Tri-alkyl organotins, biodegradation.
Abstract
This research was carried out to determine the toxicity of diphenyltin chloride on viable count of freshwater aerobic bacteria. Water samples obtained from a depth of 15 cm at five sampling points along river Benue were spiked with 3 mM of DPTCl to have a duplicate setup of three (3) treatments. Organotin degradation was determined by measuring the decrease in the concentration of DPTCl using an X-ray fluorescence spectrophotometer on days 0, 14, 28, 42, and 56 and the viable count of bacteria was also determined using spread plate technique on MSA. DPTCl concentration was further varied (5 mM, 7 mM and 10 mM) to test levels of toxicity to viable count of bacteria as well as bacterial resistance. patio sequencing technique was used to determine the metagenomics of bacteria species capable of DPTCl degradation from the water samples. Percentage of chemically degraded (40.29%) during the initial two weeks of experiment when the bacterial population was high, was more than the percentage degraded (11.49%) during the last two weeks of the experiment when the biocidal effect of DPTCl had drastically reduced the bacterial population. Of the initial bacterial viable count of 47.2 x 102 that survived the 56 days degradation exercise, only 1.8x101 (0.38%) grew on 7 mM concentration of DPTCl, and none on 10 mM DPTCl. The impact of DPTCl on the viable count of bacteria was statistically significant. Stenotrophomonas spp (29.6%) was the dominant bacterium in DPT degradation, others include; Comamonas spp (9.31%), Microbacterium spp (8.41%) Pseudomonas viridiflava (4.18%), and Stenotrophomonas geniculata (3.46%). Organotins. Diphenyltin though a di-alkyl organotin, is equally a toxic and persistent pollutant that cannot be easily degraded without the use of nutrient amendment. The toxic effect of DPT as demonstrated in this study shows that TPT and other tri-alkyl organotin should not be considered detoxified especially against microorganisms simply because they have been de-alkylated.Sites contaminated by organotins should not be considered remediated until all di and mono alkyl derivatives are completely degraded. Since our natural environments usually have no sufficient nutrient amendments for elimination of pollutants, there is need to engage abiotic processes that can assist the activities of microorganisms in order to speed up remediation process.
Introduction
Di-alkyl organotin compounds are generally used in mixtures in various amounts as heat stabilizers in the manufacture of polyvinyl chloride (PVC) and chlorinated polyvinyl chloride (CPVC) plastics. Di-alkyl tins have also been approved as PVC stabilizers for materials in contact with food and water [1]. Triphenyltins are used in agriculture as fungicides for crop protection against fungi and are directly introduced into aquatic systems via runoff from agricultural fields [2]. In addition, diphenyltins (DPTs) are known to be generated by the degradation of triphenyltin (TPT) through biological, ultraviolet irradiation, chemical, or thermal mechanisms [3]. Sewage sludge, municipal and industrial wastewater, and landfill leachates have also been discovered to constitute major sources of environmental organotins [4].
The diverse applications of di-alkyl organotins such as DPT and the eventual de-alkylation of TPT to DPT have made them to be common contaminants of aquatic habitats where they are present in appreciable amounts and have been detected in water, sediments, and biota [5]. Once these compounds become ecosystem pollutants, they may persist for long periods and how long they persist depends on the presence, amounts, and potency of biotic and abiotic factors and mechanisms needed for their remediation.
Organotin degradation is achieved by both biotic and abiotic factors. While photodecomposition by ultraviolet (UV) light is the most important abiotic degradation process, biological processes are the most important biotic factor affecting the degradation of the OTCs in aquatic and terrestrial ecosystems [6]. The degradation of organotins in the environment occurs as a progressive elimination of organic groups from Sn cations. As successive organic groups are removed, toxicity is generally reduced. Tri-alkyl organotins have been therefore considered the most toxic organotin compounds [7]. Recent findings have however shown that although tri-alkyl organotin compounds are generally considered the most toxic, there are instances where mono and di-substituted organotins are as toxic, or more toxic, than their tri-alkyl derivatives to microorganisms and other life forms [8]. Thus, agreeing with the earlier findings of Cooney [9] who reported that deacetylation in the sequence TBT --* DBT -~ MBT --* Sn, for example, does not always detoxify Triorganotins for all microorganisms. The metabolites of TPT such as DPT are therefore either more bioavailable or more toxic than the parent compound, or both [8].
Even though tri-alkyl organotins are generally considered to be more toxic, the presence of DPT in some instances has been observed to pose more threat than TPT hence the need to ensure that further degradation steps, as well as a microorganism capable of further breaking down di and monoalkyl organotins, are sought and adequately used to completely cleaning up sites contaminated by organotins. The objectives of this research therefore include:
- To assess the physicochemical and microbiological characteristics of river Benue - a freshwater habitat.
- Monitor the growth of bacteria in the presence of DPTCl and screen bacteria for their ability to resist various concentrations or DPTCl
Identify bacteria capable of degrading DPTCl from river Benue using conventional culture and molecular approaches.
Materials And Methods
During the past few decades, the runoff of river Benue has decreased markedly due to irrigation, flooding has increased contact of the river with household paints, furniture, and other materials which are usually painted with organotin formulations. Few boating activities take place in the river and series of irrigation farming also take place on the river banks where pesticides and herbicides containing organotins are used. These are possible sources of contamination of the river
Chemicals
Chemicals used both for sterilization and directly during the experiment were used without additional purity. DPTCL (96% purity) was obtained from Sigma-Aldrich chemical company Germany.
Sampling
Water samples were obtained from the surface (10 – 20 cm) at 5 points along river Benue in Makurdi. Samples were collected using 1 liter capacity plastic bottles sterilized using 90% alcohol and immediately transported to the laboratory for analysis. Standard procedures were employed to prevent contamination of samples. The collected samples were used within a week for physiochemical and bacteriological analysis
Determination of Physicochemical Parameters of Water
The following Physicochemical parameters of water were analyzed; pH, temperature, conductivity, nitrate, phosphate, Total dissolved solids (TDS), sulfate, total organic carbon (TOC), and potassium were determined using standard methods described by APHA [10].
Biodegradation Setup
The biodegradation setup was done using methods described by Ebah and Ichor [11]. Water samples obtained from the five (5) locations along river Benue were mixed and separately transferred into bowls based on the three treatments to be used. Each treatment was set up in duplicate.
The setup comprised of the following in their ratios:
Treatment A: 3.0 mM DPTCl + 20 g NPK + 1 Litre of water
Treatment B: 3.0 mM DPTCl + 20 g poultry dung + 1 Litre of water
Treatment C: 3.0 mM DPTCl + 1 Litre of water (Control)
The treatments were mixed once weekly to ensure sufficient oxygen supply to microorganisms so as to encourage aerobic activity as suggested by Burton and Turner [12]. Sampling was done on days 0, 14, 28, 42, and 56 respectively.
DPTCl degradation using the X-ray fluorescence spectrophotometer
Organotin degradation was determined by measuring the decrease in the concentration of DPTCl using an X-ray fluorescence spectrophotometer on days 0, 14, 28, 42, and 56 [11].
Determination of Viable Counts
Viable counts of organotin-utilizing bacteria were determined using the spread plate method on mineral salt agar (MSA), containing 3 mM TBTCl while the control was plated without TBTCl. Standard methods used here were similar to those described by Ebah et al., [13].
Screening of DPTCl Resistant Bacteria
The bacterial isolates which grew on MSA with 3.0 mM TBTCl were subcultured on MSA containing 5.0 mM DPTCl and further unto 7.0 mM DPTCl and finally 10.0 mM DPTCl. Isolates showing varying range of tolerance to DPTCl that is 5 mM, 7 mM and 10 mM were selected for further characterization
Identification of TBTCl Resistant Bacterial Isolates
Biochemical Identification
DPTCl utilizing bacterial Isolates was identified and characterized using the following biochemical tests; Citrate utilization, Glucose, Lactose, Mannitol, Motility, Urease test, Voges –PProskauertest, Oxidase test, Grams Reaction, Indole test, Sucrose
Molecular Analysis
The molecular analysis methodology was based on PCR and metagenomics analysis.
DNA Extraction from Water Samples
DNA extraction was done using the MoBio Power water DNA isolation Kit following the MoBio extraction, manufacturer’s instructions
Polymerase Chain Reaction Analysis
The PCR program was as follows; denaturing step at 95˚C for 3 min, followed by 33 cycles of 30 sec at 95˚C for 30 sec at 55˚C and extension at 72˚C for 1 min, followed by a final extension at 72˚C for 7 min and held at 4˚C. Amplified DNA was presented for sequencing.
DNA Sequencing
pacBio sequencing technique was used to determine the nucleotide sequence of all microorganisms present in the water sample. Sequencing of samples was based on the sequel system by pacBio Raw threads were processed through the SMRTlink (V9.0) Circular Consensus Sequences (CCS) algorithm to produce highly accurate reads (>Qv40). These highly accurate reads were processed through search and taxonomic information was determined based on QIMME2
Phylogenetics
Qualities of the DNA sequence were sequenced, and the sequences were aligned with the help of RDP (Ribosomal database project) database. Alignment was done using the multiple alignment program muscle of MEGA7 and the closest similarities were matched by using the segmatch program of RDP. After alignment with RDP database, the closely related sequences were retrieved, a distance matrix was generated and a phylogenetic tree was constructed by the neighbor joining algorithm. The stability of the phylogenetic tree was tasted with sets of 100 resamplings using bootstrap program (MEGA 7)
Results
Slight differences were observed in the physicochemical properties of water across the five (5) sampling locations. Electrical conductivity and TDS values varied significantly across the sampling points with the highest values of 631 µS/cm and 148 mg/L respectively recorded at the Wurukum sampling station. Mean EC and TDS values were 391±98.17 µS/cm and 97.8±21.0 mg/L respectively.
Nitrates and phosphates also varied relatively across the locations with mean values of 8.52±2.72 mg/L and 0.68±0.22 mg/L respectively. The two parameters had the highest values at the Wurukum sampling point. Other details of various physicochemical analyses can be seen in Table 1
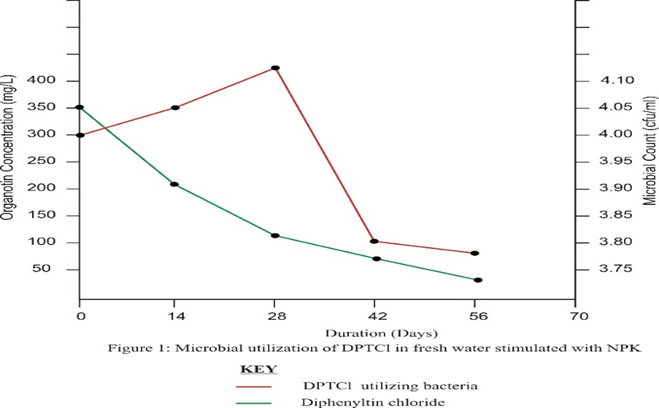
In monitoring the degradation of DPTCl using the x-ray fluorescence spectrophotometer on days 0, 14, 28, 42, and 56, it was observed that the concentration of test organotin reduced gradually while the bacteria population initially increased and then began to drop after 4 weeks. In treatment A, test organotin was reduced by 92.73 % from 350 mg/L on day 0 to 25.5 mg/L on day 56. Percentage reduction in test organotin was highest during the initial 14 days (40.29%) after which the rate of degradation dropped to a level where only 11.49% was degraded during the final 7 days of degradation.
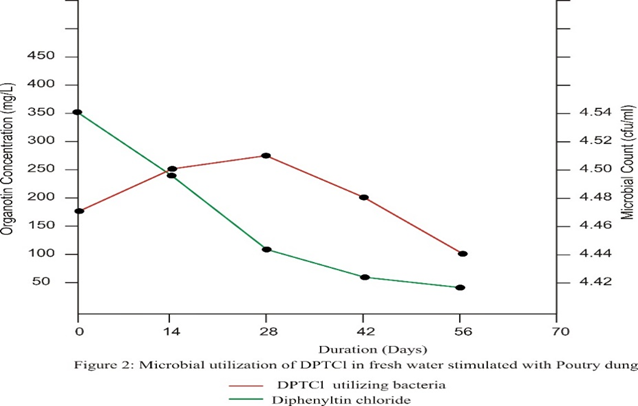
Treatment B which was amended with poultry dung, recorded a similar situation as microbial activity reduced the concentration of chemicals by 88.37% from 350mg/L on day 0 to 40.7mg/L on day 56. Although the percentage of chemical degradation seemed higher in treatment A (92.73) as compared to treatment B (88.37), this difference was statistically not significant (p>0.05)
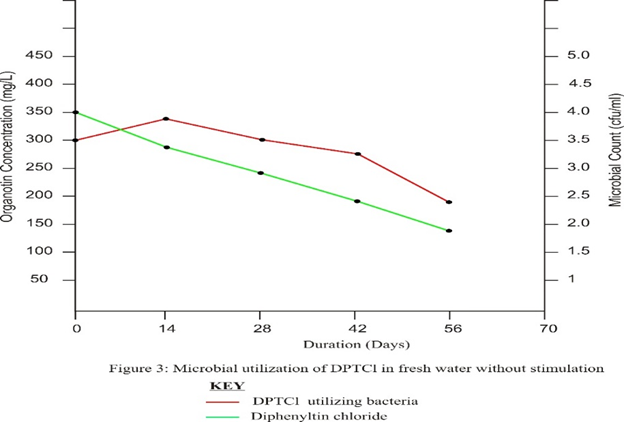
Treatment C which was a control had no nutrient amendment and recorded the least microbial growth as well as organotin degradation.
Microbes were further sub-cultured on mineral salt agar incorporated with 5mM, 7mM, and 10mM concentrations of organotin to test for microbial resistance and ability to utilize organotin. In treatment A, 2.65% of the microbial population was resistant to a 5mM concentration of DPTCl while only 0.32% was resistant to a 7mm concentration of the test chemical. In treatment B, 3.62% and 0.38% of bacteria were resistant to 5mM and 7mM concentration respectively. Only 0.30 % of bacterial population from the control sample (C) was resistant to 7Mm of test chemical. Viable count of bacteria significantly reduced with increasing concentration. In treatment A for example, of the bacteria population of 47.1x102 cfu/ml. only 1.5x101still grew on MSA+7mM and none grew on MSA+10mM DPTCl in all the treatments.
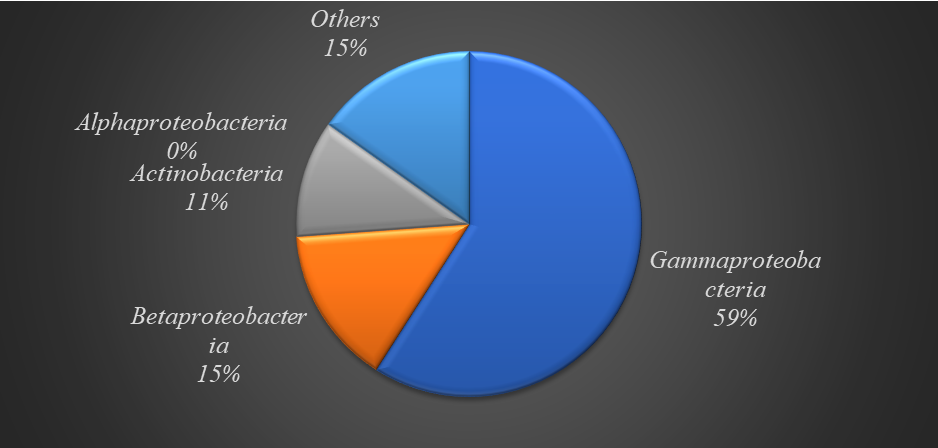
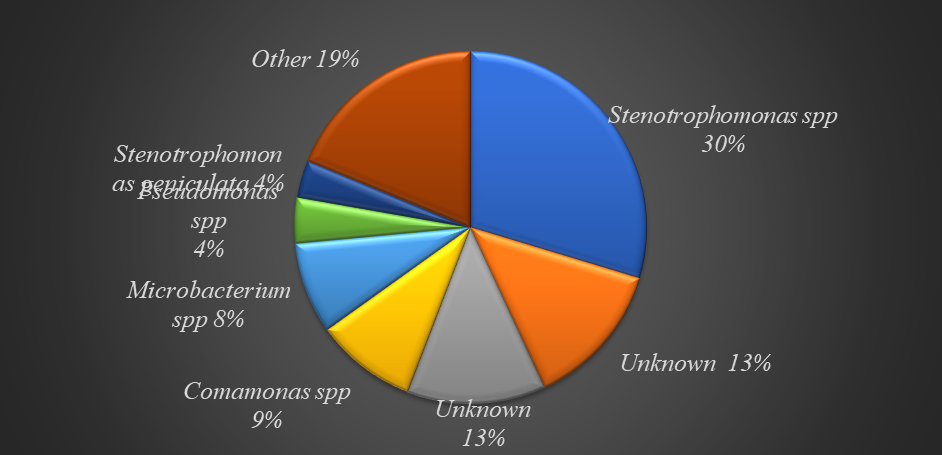
87.07% of DPTCl degraders belong to the phylum Proteobacteria. The class Gammaproteobacteria was the most culpable in DPTCl degradation and was followed by betaproteobacteria
Stenotrophomonas spp (29.6%)
was the dominant bacterium in DPT degradation, others include
; Comamonas spp (9.31%), Microbacterium spp (8.41%) Pseudomonas viridiflava (4.18%), and Stenotrophomonas geniculata (3.46%).
Other bacteria species were not identified as they had no close resemblance in NCBI database.
degradation of toxic and persistent aromatic hydrocarbons as well as organotin compounds [24].
Fig 4: Top-class classification of DPTCl degraders
Fig 5: Top Specie classification of DPTCl degraders.
Table 1. Physicochemical properties of River Benue.
| Parameter |
Wadata | Location Wurukum |
BSU |
Breweries |
Checkpoint | Mean |
| pH | 7.39 | 7.82 | 7.54 | 7.48 | 6.89 | 7.48±0.23 |
| Temperature | 27.50 | 26.80 | 27.2 | 26.0 | 28.9 | 27.3±0.76 |
| EC | 305 | 631 | 326 | 380 | 314 | 391±98.17 |
| TDS | 82 | 148 | 74 | 98 | 87 | 97.8±21.0 |
| Nitrates | 8.7 | 14.9 | 5.8 | 7.7 | 5.5 | 8.52±2.72 |
| Phosphates | 0.45 | 1.20 | 0.68 | 0.56 | 0.49 | 0.68±0.22 |
| DO | 5.20 | 4.90 | 4.70 | 4.55 | 5.08 | 4.89±0.19 |
Table 2: Screening bacteria for organotin utilization
| Treatment | MSA only | MSA +5 mM | MSA+ 7 mM | MSA + 10mM |
| A | 47.1x102 | 12.5x101 | 1.5x101 | Nil |
| B | 47.2x102 | 17.1x101 | 1.8x101 | Nil |
| C | 36.6x102 | 9.9x101 | 1.1x101 | Nil |
Table 3: Percentage (%) of Bacteria resistance to DPTCl
| Concentration | Treatment A | Treatment B | Treatment C |
| 5mM | 2.65 | 3.62 | 3.21 |
| 7mM | 0.32 | 0.38 | 0.30 |
Discussion
The physicochemistry of the freshwater ecosystem is one of the major factors that determine the existence and distribution of all aquatic life. This study further revealed that the presence, growth, and distribution of the microbial population to a large extent is determined by factors such as pH, Nitrates, Phosphates, Dissolved oxygen, and electrical conductivity.
The Wurukum sampling station recorded the highest values of physicochemical parameters. For example, nitrate and phosphate values of 14.9 mg/L and 1.20 mg/L respectively were recorded. This location is characterized by massive irrigation farming, as well as multiple human activities. Waste generated from an abattoir that is sited here is another factor considered to be responsible for the high nitrate and phosphate levels reported here. Mean Nitrate values of all samples obtained from the various locations was 8.52±3.40 and though within the maximum limits specified by WHO (2011) of 50mg/L, there is an obvious increase in their concentrations when compared to previous research. This research agrees with the findings of Kondum et al., [15] in the same study area, who reported nitrate levels of between 8.34±0.30 and 14.69±1.35 mg/L. The nitrate values here however vary with the earlier findings of Okenyi et al., [16] who reported very low values of 2.0 and 2.7 mg/L. This shows that in recent years, eutrophication has increased the concentrations of these compounds in our rivers.
Investigated physicochemical parameters indicate that river Benue-a freshwater habitat is slightly polluted and when compared with previous findings, it is observed that over the years, eutrophication has led to an increase in nitrates and phosphates levels while depleting the dissolved oxygen.
The high rate of degradation during the initial fourteen days of the experiment could be a result of the interplay of both biotic and abiotic factors. The high initial bacteria population combined with the effect of sunlight increased the uptake and degradation of DPTCl. The impact of this collaboration was however short-lived as some bacteria could not stand the biocidal effect of the test chemical. This subsequently reduced rate at which the chemical was degraded during the later stages of the experiment.
There is evidence to show that microorganisms can slowly degrade TPT into inorganic tin via di- and monophenyltins. Microbial interactions with tin are important because microbes are at the base of the food web and because they have the potential for bioremediation [17] [18][7]. The findings here also prove this as the percentage of chemical degradation (40.29%) during the initial two weeks of the experiment when the bacterial population was high, is more than the percentage degraded (11.49%) during the later stages of the experiment when the biocidal effect of the chemical had drastically reduced bacterial population.
92.73 % of DPTCl was degraded by bacteria leaving behind only 7.27 % of test chemicals after 56 days of degradation. Reduction of TBTCl (a tri alkyl organotin) to as low as 0.98
References
- European Food Safety Authority (EFSA) (2004). Scientific panel on contaminants in the food chain, opinion on the health risks assessment to consumers associated with the exposure to organotins in foodstuffs. Question no EFSA-2003 – 110, EFSA Journal 102: 1-119.
View at Publisher | View at Google Scholar - Fent, K. (1996). Organotin compounds in municipal wastewater and sewage sludge: contamination, fate in treatment process and ecotoxicological consequences. Science of the Total Environment, 185(1-3), 151-159.
View at Publisher | View at Google Scholar - WHO-IPCS. (1999). World health organization. The international programme on chemical safety. Tributyl compounds. Environ Health Criteria 116.
View at Publisher | View at Google Scholar - Santos, D. M., Araújo, I. P., Machado, E. C., Carvalho-Filho, M. A., Fernandez, M. A., Marchi, M. R., & Godoi, A. F. L. (2009). Organotin compounds in the Paranaguá Estuarine Complex, Paraná, Brazil: Evaluation of biological effects, surface sediment, and suspended particulate matter. Marine Pollution Bulletin, 58(12), 1926-1931.
View at Publisher | View at Google Scholar - Bellas, J., Hilvarsson, A., & Granmo, Å. (2005). Sublethal effects of a new antifouling candidate on lumpfish (Cyclopterus lumpus L.) and Atlantic cod (Gadus morhua L.) larvae. Biofouling, 21(3-4), 207-216.
View at Publisher | View at Google Scholar - de Carvalho Oliveira, R., & Santelli, R. E. (2010). Occurrence and chemical speciation analysis of organotin compounds in the environment: a review. Talanta, 82(1), 9-24.
View at Publisher | View at Google Scholar - Okoro, H. K., Fatoki, O. S., Adekola, F. A., Ximba, B. J., & Snyman, R. G. (2011). Sources, Environmental levels and toxicity of organotin in marine environment–a review.
View at Publisher | View at Google Scholar - Paton, G. I., Cheewasedtham, W., Marr, I. L., & Dawson, J. J. C. (2006). Degradation and toxicity of phenyl tin compounds in soil. Environmental Pollution, 144(3), 746-751.
View at Publisher | View at Google Scholar - Cooney, J. J. (1995). Organotin compounds and aquatic bacteria: a review. Helgoländer Meeresuntersuchungen, 49, 663-677.
View at Publisher | View at Google Scholar - APHA AWWA, W. P. C. F. (1998). Standard Methods for the Examination of water and Wastewater 20th edition. American Public Health Association, American Water Work Association, Water Environment Federation, Washington, DC.
View at Publisher | View at Google Scholar - Ebah, E. E., & Ichor, T. (2018). Aerobic Degradation of Tributyltin Chloride and Dip-Henyltin Chloride by Marine Bacteria from Onne Port Nigeria. J Environ Anal Toxicol, 8(582), 2161-0525.
View at Publisher | View at Google Scholar - Burton, C. H., & Turner, C. (2003). Manure management: Treatment strategies for sustainable agriculture. Editions Quae.
View at Publisher | View at Google Scholar - Ebah, E., Ichor, T., & Okpokwasili, G. C. (2016). Isolation and biological characterization of tributyltin degrading bacteria from onne port sediment. Open Journal of Marine Science, 6(2), 193-199.
View at Publisher | View at Google Scholar - World Health Organization. (2004). Guidelines for drinking-water quality (Vol. 1). World Health Organization.
View at Publisher | View at Google Scholar - Kondum, F. A., Iwar, R. T., & Kon, E. T. (2021). A comparison of water quality indexes for an inland river. Journal of Engineering Research and Reports, 20(4), 1-14.
View at Publisher | View at Google Scholar - Okenyi, B.E., Ngozi-Olehi, L.C. and Ibe C. O. (2016). Physico-Chemical Index of River Benue and Its Implications Journal of Applied Chemistry (9)4: 53-55 e-ISSN: 2278-5736.
View at Publisher | View at Google Scholar - Inoue, H., Takimura, O., Fuse, H., Murakami, K., Kamimura, K., & Yamaoka, Y. (2000). Degradation of triphenyltin by a fluorescent pseudomonad. Applied and environmental microbiology, 66(8), 3492-3498.
View at Publisher | View at Google Scholar - Gadd, G. M. (2000). Microbial interactions with tributyltin compounds: detoxification, accumulation, and environmental fate. Science of the total environment, 258(1-2), 119-127.
View at Publisher | View at Google Scholar - White, J. S., Tobin, J. M., & Cooney, J. J. (1999). Organotin compounds and their interactions with microorganisms. Canadian journal of microbiology, 45(7), 541-554.
View at Publisher | View at Google Scholar - Su, G., Zhang, X., Raine, J. C., Xing, L., Higley, E., Hecker, M., ... & Yu, H. (2013). Mechanisms of toxicity of triphenyltin chloride (TPTC) determined by a live cell reporter array. Environmental Science and Pollution Research, 20, 803-811.
View at Publisher | View at Google Scholar - Bhowmik, S. N., & Singh, C. S. (2004). Mass multiplication of AM inoculum: effect of plant growth-promoting rhizobacteria and yeast in rapid culturing of Glomus mosseae. Current Science, 705-709.
View at Publisher | View at Google Scholar - Gao, J., Ye, J., Ma, J., Tang, L., & Huang, J. (2014). Biosorption and biodegradation of triphenyltin by Stenotrophomonas maltophilia and their influence on cellular metabolism. Journal of hazardous materials, 276, 112-119.
View at Publisher | View at Google Scholar - Thatheyus, A. J., & Selvam, A. G. (2013). Synthetic pyrethroids: toxicity and biodegradation. Appl Ecol Environ Sci, 1(3), 33-36.
View at Publisher | View at Google Scholar - Chen, Y. X., Liu, H. E., & Chen, H. L. (2003). Characterization of phenol biodegradation by Comamonas testosteroni ZD4-1 and Pseudomonas aeruginosa ZD4-3. Biomedical and environmental sciences: BES, 16(2), 163-172.
View at Publisher | View at Google Scholar